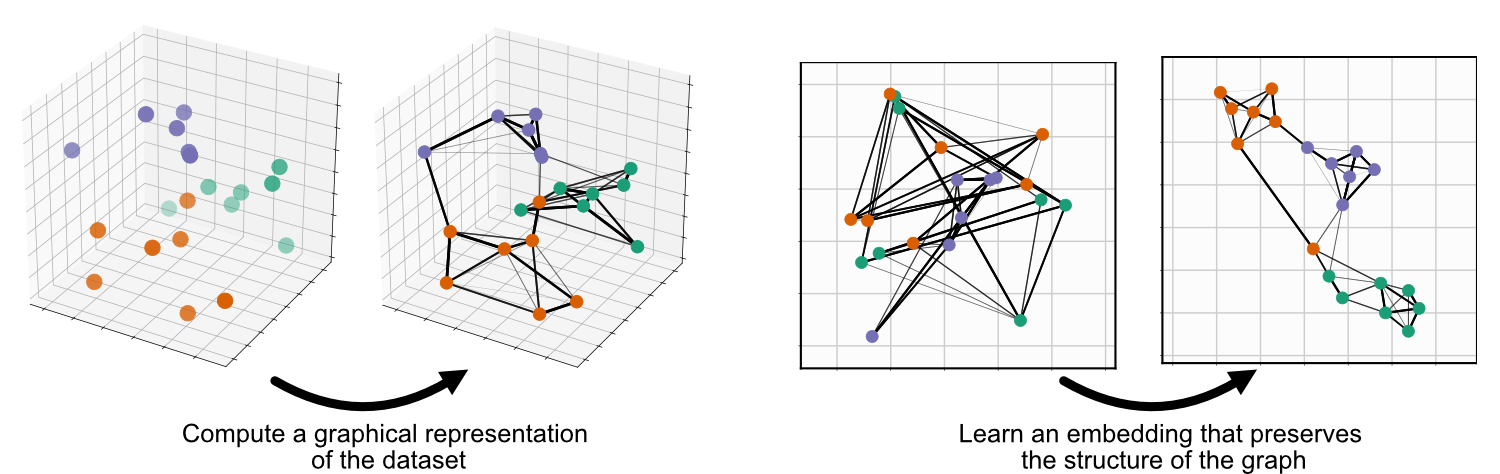
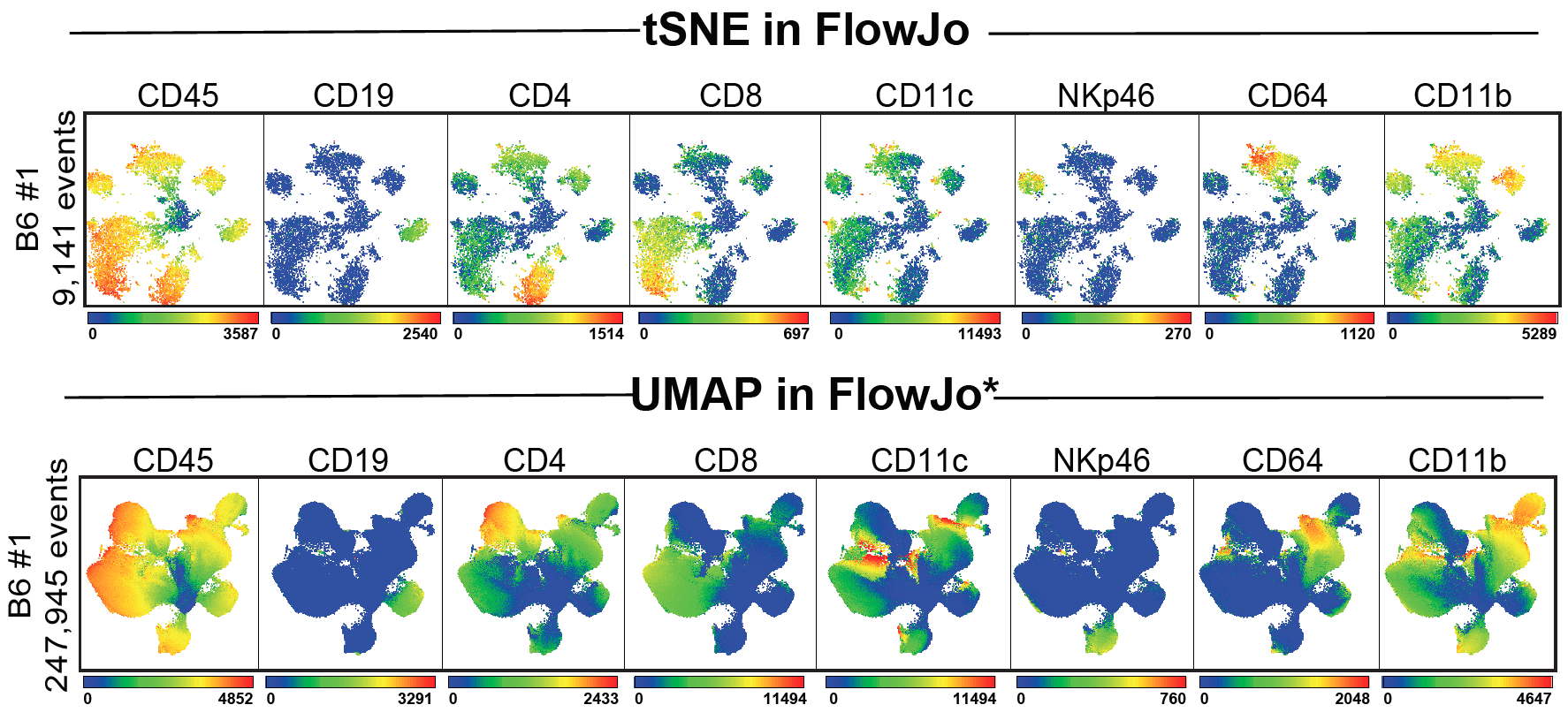
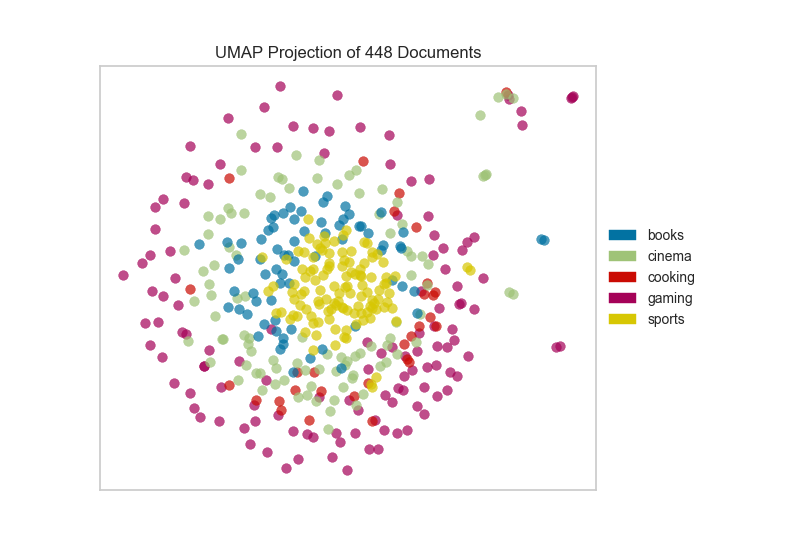
Umap Init. As the number of data points increase, UMAP becomes more time efficient compared to TSNE. One of the most widely used techniques for visualization is t-SNE, but its performance suffers with large datasets and using it correctly can be challenging. UMAP is a general purpose manifold learning and dimension reduction algorithm. It seeks to learn the manifold structure of your data and find a low dimensional embedding that preserves the essential topological structure of that manifold. latest User Guide / Tutorial: How to Use UMAP Basic UMAP Parameters Plotting UMAP results UMAP Reproducibility Transforming New Data with UMAP Inverse transforms Parametric (neural network) Embedding UMAP on sparse data UMAP for Supervised Dimension Reduction and Metric Learning Using UMAP for Clustering Outlier detection using UMAP UMAP, like t-SNE, can also create tears in clusters that are not actually present, resulting in a finer clustering than is necessarily present in the data. The default string "spectral" computes an initial embedding using eigenvectors of the connectivity graph matrix. If you are already familiar with sklearn you should be able to use UMAP as a drop in replacement for t-SNE and other dimension reduction classes.

Umap Init. One of the most widely used techniques for visualization is t-SNE, but its performance suffers with large datasets and using it correctly can be challenging. Release of the UnrealEd software program prompted the development of the Unreal Engine Map File file type by Epic Games. It is designed to be compatible with scikit-learn, making use of the same API and able to be added to sklearn pipelines. UMAP is a nonparametric graph-based dimensionality reduction algorithm using applied Riemannian geometry and algebraic topology to find low-dimensional embeddings of structured data. The default string "spectral" computes an initial embedding using eigenvectors of the connectivity graph matrix. An unordered_map can be initialized in different ways like: simple initialization using assignment operator and subscript operator. initializing using a Initializer List. initializing using pair of arrays. initializing from another map. simple initialization using assignment operator and subscript operator. Umap Init.
UMAP is a nonparametric graph-based dimensionality reduction algorithm using applied Riemannian geometry and algebraic topology to find low-dimensional embeddings of structured data.
UMAP is a general purpose manifold learning and dimension reduction algorithm.
Umap Init. Some of the following help text is lifted verbatim from the Python reference implementation at https://github.com/lmcinnes/umap. The default string "spectral" computes an initial embedding using eigenvectors of the connectivity graph matrix. UPK Unreal Package such as objects, light brightness and direction, fog density and color, and. If you are already familiar with sklearn you should be able to use UMAP as a drop in replacement for t-SNE and other dimension reduction classes. Description. –list [charset] Without argument list all character sets recognized. It seeks to learn the manifold structure of your data and find a low dimensional embedding that preserves the essential topological structure of that manifold. latest User Guide / Tutorial: How to Use UMAP Basic UMAP Parameters Plotting UMAP results UMAP Reproducibility Transforming New Data with UMAP Inverse transforms Parametric (neural network) Embedding UMAP on sparse data UMAP for Supervised Dimension Reduction and Metric Learning Using UMAP for Clustering Outlier detection using UMAP UMAP, like t-SNE, can also create tears in clusters that are not actually present, resulting in a finer clustering than is necessarily present in the data.
Umap Init.